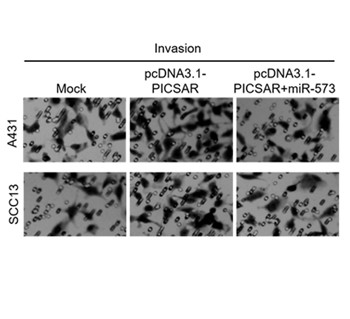
Retracted: MicroRNA-573 inhibits cell proliferation, migration and invasion and is downregulated by PICSAR in cutaneous squamous cell carcinoma
DOI:
https://doi.org/10.17305/bjbms.2021.6301Keywords:
MicroRNA-573, Long non-coding RNA PICSAR, Cutaneous squamous cell carcinoma, Proliferation, Migration, InvasionAbstract
This article has been retracted: https://doi.org/10.17305/bjbms.2022.8232
Retraction: "MicroRNA-573 functions as a tumor suppressor and is downregulated by PICSAR in cutaneous squamous cell carcinoma" by Yanhua Wang, Shengjian Tang and Jianping Lv, Bosn J Basic Med Sci. 2022 Apr 1;22(2):229-237. (doi: 10.17305/bjbms.2021.6301). This article has been withdrawn at the request of the authors. The authors voluntarily requested the retraction of this paper. The additional experiments they conducted could not confirm the role of microRNA-573 functions in cutaneous squamous cell carcinoma reported in the BJBMS article (vol. 22(2):229-237). All authors signed and agreed on the retraction of the article. The Bosnian Journal of Basic Medical Sciences apologizes to the readers and scientific community for the inconveniences caused by this retraction.
Citations
Downloads
Downloads
Published
Issue
Section
Categories
License
Copyright (c) 2021 Yanhua Wang, Shengjian Tang, Jianping Lv

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2021-09-16
Published 2022-04-01









