Pharmacogenomics biomarkers for personalized methadone maintenance treatment: The mechanism and its potential use
DOI:
https://doi.org/10.17305/bjbms.2020.4897Keywords:
Methadone, pharmacokinetics, pharmacogenomics, personalized medicine, ABCB1, CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP2D6Abstract
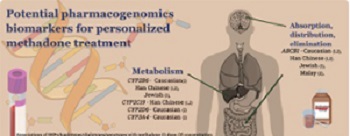
Methadone has a wide pharmacokinetic interindividual variability, resulting in unpredicted treatment response. Pharmacogenomic biomarkers seem promising for personalized methadone maintenance treatment. The evidence supports the use of ABCB1 single-nucleotide polymorphism (SNP) 1236C>T with genotypes C/T or C/C (Jewish) and haplotypes AGCTT carrier, AGCGC heterozygote, or non-carrier (Caucasian), which have a predicted lower methadone dose requirement. In contrast, ABCB1 SNP 1236C>T with genotype T/T (Jewish); haplotypes AGCGC homozygote, AGCTT non-carrier (Caucasian), and ABCB1 3435C>T variant carrier; and haplotypes CGT, TTC, and TGT (Han Chinese) have a predicted higher methadone dose. For methadone plasma levels, ABCB1 diplotype non-CGC/TTT (Malay) predicted lower, and diplotype CGC/TTT (Malay), 3435C>T allelic carrier, haplotypes (CGT, TTC, TGT) (Han Chinese) predicted higher methadone levels. In terms of metabolism biomarkers, a lower methadone requirement was related to carriers of CYP2B6 genotypes *4(G/G) and *9(T/T) among Jewish patients, CYP2B6*9 genotype (T/T) and haplotypes (TA/TG); and CYP2C19 (*2/*2,*2/*3, and *3/*3; Han Chinese). Higher methadone dose was observed in CYP2C19*1 allelic carriers (Han Chinese) and CYP2D6 ultrarapid metabolizer (Caucasian). Lower methadone levels were reported in CYP2B6 SNPs, haplotypes TTT, and AGATAA (Han Chinese), CYP2C19 genotype *1/*1 (Han Chinese), allelic carrier *1xN (Caucasian), and CYP3A4 genotype *1/*1 (Caucasian). Carriers of CYP2B6 genotype *6/*6 (Caucasian), CYP2B6 haplotypes ATGCAG and ATGCTG (Han Chinese), and CYP3A4 genotype *1/*1B (Caucasian) had predicted higher methadone plasma levels. Specific pharmacokinetics biomarkers have potential uses for personalized methadone treatment in specific populations.
Citations
Downloads
Downloads
Additional Files
Published
How to Cite
Accepted 2020-08-10
Published 2021-04-01









