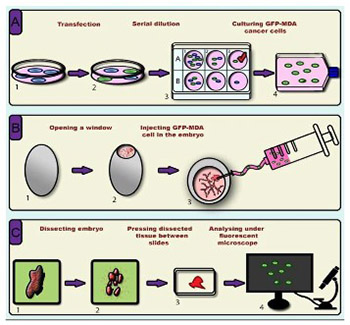
A novel in ovo model to study cancer metastasis using chicken embryos and GFP expressing cancer cells
DOI:
https://doi.org/10.17305/bjbms.2019.4372Keywords:
Tumor invasion, in ovo models, chicken embryo model, metastasis, GFP cellsAbstract
Cancer metastasis is the leading cause of cancer-related mortality worldwide. To date, several in vitro methodologies have been developed to understand the mechanisms of cancer metastasis and to screen various therapeutic agents against it. Nevertheless, mimicking an in vivo microenvironment in vitro is not possible; while in vivo experiments are complex, expensive and bound with several regulatory requirements. Herein, we report a novel in ovo model that relies on chicken embryo to investigate cancer cell invasion and metastasis to various organs of the body. In this model, we directly injected green fluorescent protein (GFP) expressing cancer cells to the heart of chicken embryo at 3 days of incubation, then monitored cell migration to various organs. To this end, we used a simple tissue processing technique to achieve rapid imaging and quantification of invasive cells. We were able to clearly observe the migration of GFP expressing cancer cells into various organs of chicken embryo. Organ specific variation in cell migration was also observed. Our new slide pressing based tissue processing technique improved the detectability of migrated cells. We herein demonstrate that the use of GFP expressing cancer cells allows easy detection and quantification of migrated cancer cells in the chicken embryo model, which minimizes the time and effort required in this types of studies compared to conventional histopathological analysis. In conclusion, our investigation provides a new cancer metastasis model that can be further improved to include more complex aspects, such as the use of multiple cell lines and anti-metastatic agents, thus opening new horizons in cancer biology and pharmaceutical research.
Citations
Downloads