HLA genotyping in pediatric celiac disease patients
DOI:
https://doi.org/10.17305/bjbms.2014.3.28Keywords:
celiac disease, HLA-DQ alleles, HLA genotypingAbstract
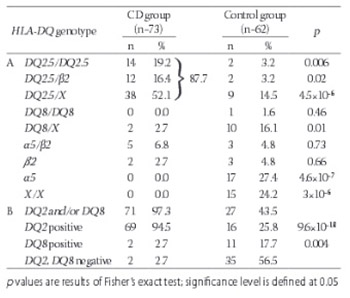
Celiac disease (CD) is a chronic inflammatory disease in the small intestine triggered by gluten uptake that occurs in genetically susceptible individuals. HLA-DQ2 protein encoded by HLA-DQA1*05 and DQB1*02 alleles is found in 90-95% of CD patients. All of the remaining patients carry HLA-DQ8 protein encoded by HLA-DQA1*03 and DQB1*03:02 alleles. Specific HLA-DQ genotypes define different risk for CD incidence. Presence of susceptible HLA-DQ genotypes does not predict certain disease development, but their absence makes CD very unlikely, close to 100%. Here we presented for the first time the distribution of HLA-DQ genotypes in the group of pediatric celiac patients from the University Children’s Hospital, Belgrade, Serbia and estimated risk for CD development that these genotypes confer. Seventy three celiac disease patients and 62 healthy individuals underwent genotyping for DQA1, DQB alleles and DRB1 allele. 94.5% of patients carried alleles that encode DQ2 protein variant and 2.7% carried alleles that encode DQ8 protein variant. Two patients carried single DQB1*02 allele. No patients were negative for all the alleles predisposing to CD. The highest HLA-DQ genotype risk for CD development was found in group of patients homozygous for DQ2.5 haplotype, followed by the group of heterozygous carriers of DQ2.5 haplotype in combination with DQB1*02 allele within the other haplotype. The lowest risk was observed in carriers of a single copy of DQB1*02 or DQA1*05 allele or other non-predisposing alleles. HLA genotyping, more informative than serological testing commonly used, proved to be a useful diagnostic tool for excluding CD development.
Citations
Downloads
References
Fasano A, Berti I, Gerarduzzi T, Not T, Colletti RB, Drago S, et al. Prevalence of celiac disease in at-risk and not-at-risk groups in the United States: a large multicenter study. Arch Intern Med 2003; 163(3): 286-292.
Dubé C, Rostom A, Sy R, Cranney A, Saloojee N, Garritty C, et al. The prevalence of celiac disease in average-risk and at-risk Western European populations: a systematic review. Gastroenterology 2005; 128(4 Suppl 1): S57-67.
Mustalahti K, Catassi C, Reunanen A, Fabiani E, Heier M, McMillan S, et al. The prevalence of celiac disease in Europe: results of a centralized, international mass screening project. Ann Med 2010; 42(8): 587-595.
Sanders DS, Hurlstone DP, Stokes RO, Rashid F, Milford-Ward A, Hadjivassiliou M, et al. Changing face of adult coeliac disease: experience of a single university hospital in South Yorkshire. Postgrad Med J 2002; 78(915): 31-33.
van Heel DA, West J. Recent advances in coeliac disease. Gut 2006; 55(7): 1037–1046.
Leeds JS, Hopper AD, Sanders DS. Coeliac disease. Br Med Bull 2008; 88 (1): 157-170.
Llorente-Alonso MJ, Fernández-Acenero MJ, Sebastián M. Gluten intolerance: Sex-and age-related features. Can J Gastroenterol 2006; 20(11): 719-722.
Megiorni F, Mora B, Bonamico M, Barbato M, Montuori M, Viola F, et al. HLA-DQ and susceptibility to celiac disease: evidence for gender differences and parent-of-origin effects. Am J Gastroenterol 2008; 103(4): 997-1003.
Esteve M, Rosinach M, Fernández-Bañares F, Farré C, Salas A, Alsina M, et al. Spectrum of gluten-sensitive enteropathy in first-degree relatives of patients with coeliac disease: clinical relevance of lymphocytic enteritis. Gut 2006; 55(12): 1739–1745.
Greco L, Romino R, Coto I, Di Cosmo N, Percopo S, Maglio M, et al. The first large population based twin study of coeliac disease. Gut 2002; 50(5): 624-628.
Lionetti E, Catassi C. New clues in celiac disease epidemiology, pathogenesis, clinical manifestations, and treatment. Int Rev Immunol 2011; 30(4): 219–231.
Matysiak-Budnik T, Moura IC, Arcos-Fajardo M, Lebreton C, Ménard S, Candalh C, et al. Secretory IgA mediates retrotranscytosis of intact gliadin peptides via the transferrin receptor in celiac disease. J Exp Med 2008; 205(1): 143-154.
Caillat-Zucman S. Molecular mechanisms of HLA association with autoimmune diseases. Tissue Antigens 2009; 73(1): 1-8.
Karell K, Louka AS, Moodie SJ, Ascher H, Clot F, Greco L, et al. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol 2003; 64(4): 469-477.
Margaritte-Jeannin P, Babron MC, Bourgey M, Louka AS, Clot F, Percopo S, et al. HLA-DQ relative risks for coeliac disease in European populations: a study of the European Genetics Cluster on Coeliac Disease. Tissue Antigens 2004; 63(6): 562-567.
Clerget-Darpoux F, Bouguerra F, Kastally R, Semana G, Babron MC, Debbabi A, et al. High risk genotypes for celiac disease. C R Acad Sci III 1994; 317(10): 931-936.
Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ, et al. The HLA-DQ2 gene dose effect in coeliac disease is directly related to the magnitude and breadth of the gluten-specific T cell responses. Proc Natl Acad Sci USA 2003; 100(21): 12390–12395.
Al-Toma A, Goerres MS, Meijer JW, Peña AS, Crusius JB, Mulder CJ. Human leukocyte antigen-DQ2 homozygosity and the development of refractory celiac disease and enteropathy-associated T –cell lymphoma. Clin Gastroenterol Hepatol 2006; 4(3): 315-319.
Green P, Cellier C. Celiac disease. N Engl J Med 2007; 357(17): 1731-1743.
Kaukinen K, Partanen J, Mäki M, Collin P. HLA-DQ typing in the diagnosis of celiac disease. Am J Gastroenterol 2002; 97(3): 695-699.
Wolters VM, Wijmenga C. Genetic background of celiac disease and its clinical implications. Am J Gastroenterol 2008; 103(1): 190-195.
Walker-Smith JA, Guandalini S, Schmitz J, Shmerling DH, Visakorpi JK. Revised criteria for diagnosis of coeliac disease. Report of working group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child 1990; 65(8): 909-911.
Lavant EH, Agardh DJ, Nilsson A, Carlson JA. A new PCR-SSP method for HLA DR-DQ risk assessment for celiac disease. Clin Chim Acta 2011; 412(9-10): 782-784.
Allele Frequency Net Database (http://www.allelefrequencies.net) [accessed: 20 February 2014]
National Marrow Donor Program Bioinformatics
(http://bioinformatics.nmdp.org/HLA/HLA_Resources.aspx) [accessed: 18 February 2014]
Megiorni F, Mora B, Bonamico M, Barbato M, Nenna R, Maiella G, et al. HLA-DQ and risk gradient for celiac disease. Hum Immunol 2009; 70(1): 55-59.
Piccini B, Vascotto M, Serracca L, Luddi A, Margollicci MA, Balestri P, et al. HLA-DQ typing in the diagnostic algorithm of celiac disease. Rev Esp Enferm Dig 2012; 104(5): 248-254.
Louka AS, Moodie SJ, Karell K, Bolognesi E, Ascher H, Greco L, et al. European Genetics Cluster on Celiac Disease. A collaborative European search for non-DQA1*05-DQB1*02 celiac disease loci on HLA-DR3 haplotypes: analysis of transmission from homozygous parents. Hum Immunol 2003; 64(3): 350-358.
Louka AS, Lie BA, Talseth B, Ascher H, Ek J, Gudjónsdóttir AH, et al. Coeliac disease patients carry conserved HLA-DR3-DQ2 haplotypes revealed by association of TNF alleles. Immunogenetics 2003; 55(5): 339-343.
Bolognesi E, Karell K, Percopo S, Coto I, Greco L, Mantovani V, et al. Additional factor in some HLA DR3/DQ2 haplotypes confers a fourfold increased genetic risk of celiac disease. Tissue Antigens 2003; 61(4): 308-316.
Liu E, Lee HS, Aronsson CA, Hagopian WA, Koletzko S, Rewers MJ, et al. Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med. 2014; 371(1): 42-9.
Vojvodić S, Ademović-Sazdanić D. HLA II class antigens and susceptibility to coeliac disease. Genetika 2011; 43(3): 517-526.
Žunec R, Grubić Z, Jurčić Z, Peršić M, Kaštelan A, Kerhin-Brkljačić V. Hla-DQ2 heterodimer in the diagnosis of celiac disease. Biochemia Medica 2004; 14: 3-4.
Dolinšek J, Micetik-turk D, Urlep-Žužej D, Zagradišnik B, Haimilia K, Holopainen P. Importance of Celiac Disease Patients Lacking Hla Dq2 or Dq8 Heterodimer in Slovenia. J Pediatr Gastroenterol Nutr 2006; 42(5): 18-19.
Mubarak A, Spierings E, Wolters V, van Hoogstraten I, Kneepkens CM, Houwen R. Human Leukocyte Antigen DQ2.2 and Celiac Disease. J Pediatr Gastroenterol Nutr 2013; 56(4): 428-430.
Sollid LM, Lie BA. Celiac disease genetics: current concepts and practical applications. Clin Gastroenterol Hepatol 2005; 3(9): 843-851.
Garner CP, Murray JA, Ding YC, Tien Z, van Heel DA, Neuhausen SL. Replication of celiac disease UK genome-wide association study results in a US population. Hum Mol Genet 2009; 18(21): 4219–4225.
Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet 2008; 40(4): 395–402.
Romanos J, van Diemen CC, Nolte IM, Trynka G, Zhernakova A, Fu J, et al. Analysis of HLA and non-HLA alleles can identify individuals at high risk for celiac disease. Gastroenterology 2009; 137(3): 834–840.
Megiorni F, Pizzuti A. HLA-DQA1 and HLA-DQB1 in Celiac disease predisposition: practical implications of the HLA molecular typing. J Biomed Sci. 2012; 19:88.
Downloads
Additional Files
Published
Issue
Section
Categories
License
Copyright (c) 2015 Bosnian Journal of Basic Medical Sciences

This work is licensed under a Creative Commons Attribution 4.0 International License.
How to Cite
Accepted 2014-07-21
Published 2014-08-16









