A comparative analysis of the effectiveness of cytogenetic and molecular genetic methods in the detection of Down syndrome
DOI:
https://doi.org/10.17305/bjbms.2014.2271Keywords:
Down syndrome, cytogenetic analysis, STR markers, QF PCRAbstract
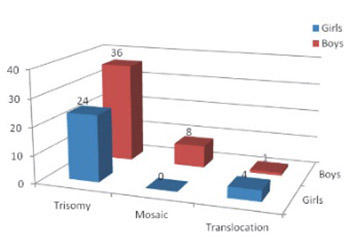
The goal of this study was to examine the effectiveness of 6 STR markers application (D21S1435, D21S11, D21S1270, D21S1411, D21S226 and IFNAR) in molecular genetic diagnostics of Down syndrome (DS) and to compare it with cytogenetic method. Testing was performed on 73 children, with the previously cytogenetically confirmed Down syndrome. DNA isolated from the buccal swab was used. Previously mentioned loci located on chromosome 21 were simultaneously amplified using quantitative fluorescence PCR (QF PCR). Using this method, 60 previously cytogenetically diagnosed DS with standard type of trisomy 21 were confirmed. Furthermore, six of eight children with mosaic type of DS were detected. Two false negative results for mosaic type of DS were obtained. Finally, five children with the translocation type of Down syndrome were also confirmed with this molecular test. In conclusion, molecular genetic analysis of STR loci is fast, cheap and simple method that could be used in detection of DS. Regarding possible false results detected for certain number of mosaic types, cytogeneticanalysis should be used as a confirmatory test.
Citations
Downloads
Downloads
Additional Files
Published
How to Cite
Accepted 2017-07-12
Published 2014-05-20









