mRNA expression of nuclear factor of activated T-cells, cytoplasmic 2 (NFATc2) and peroxisome proliferator-activated receptor gamma (PPARG) transcription factors in colorectal carcinoma
DOI:
https://doi.org/10.17305/bjbms.2017.1886Keywords:
NFATc2, PPARG, colorectal cancer, gene expression, biomarker, CRC, targeted therapyAbstract
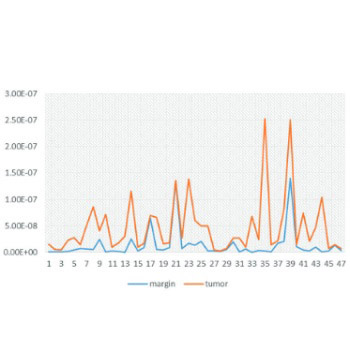
Transcription factors are involved in cell cycle and apoptosis regulation and thus have a key role in the carcinogenesis of different tumors. Nuclear factor of activated T-cells, cytoplasmic 2 (NFATc2) and peroxisome proliferator-activated receptor gamma (PPARG) transcription factors are important in the carcinogenesis of colorectal cancer (CRC). In this study, we examined whether the expression of NFATc2 and PPARG genes is significantly altered during the carcinogenesis of CRC. A total of 47 tumor samples and matched normal tissue margins were collected during surgery from patients with CRC. In addition, three CRC cell lines (HCT119, SW480, and HT29) and healthy cell line were used. After total RNA extraction and cDNA synthesis, mRNA expression levels of NFATc2 and PPARG were examined by real-time polymerase chain reaction. The results showed that NFATc2 is overexpressed in the tumor tissues compared with normal tissue margins (p ≤ 0.05). However, the mRNA expression levels of PPARG were not significantly different between the tumor tissues and tissue margins. Our results indicate that NFATc2 may be used as an early diagnostic or predictive biomarker for CRC as well as a therapeutic target, providing that upcoming studies confirm these results.
Citations
Downloads
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61(2):69-90. https://doi.org/10.3322/caac.20107.
Haggar FA, Boushey RP. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 2009;22(4):191-7.
https://doi.org/10.1055/s-0029-1242458.
Sabatino L, Pancione M, Votino C, Colangelo T, Lupo A, Novellino E, et al. Emerging role of the β-catenin-PPARγ axis in the pathogenesis of colorectal cancer. World J Gastroenterol 2014;20(23):7137-51. https://doi.org/10.3748/wjg.v20.i23.7137.
Takebe N, Ivy SP. Controversies in cancer stem cells: Targeting embryonic signaling pathways. Clin Cancer Res 2010;16(12):3106-12.
https://doi.org/10.1158/1078-0432.CCR-09-2934.
Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol 2011;8(2):97-106. https://doi.org/10.1038/nrclinonc.2010.196.
Adesina AM, Lopez-Terrada D, Wong KK, Gunaratne P, Nguyen Y, Pulliam J, et al. Gene expression profiling reveals signatures characterizing histologic subtypes of hepatoblastoma and global deregulation in cell growth and survival pathways. Hum Pathol 2009;40(6):843-53. https://doi.org/10.1016/j.humpath.2008.10.022.
de La Coste A, Romagnolo B, Billuart P, Renard CA, Buendia MA, Soubrane O, et al. Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci U S A 1998;95(15):8847-51. https://doi.org/10.1073/pnas.95.15.8847.
Kim MS, Kim SS, Ahn CH, Yoo NJ, Lee SH. Frameshift mutations of Wnt pathway genes AXIN2 and TCF7L2 in gastric carcinomas with high microsatellite instability. Hum Pathol 2009;40(1):58-64. https://doi.org/10.1016/j.humpath.2008.06.006.
Koesters R, Ridder R, Kopp-Schneider A, Betts D, Adams V, Niggli F, et al. Mutational activation of the β-catenin proto-oncogene is a common event in the development of Wilms' tumors. Cancer Res 1999;59(16):3880-2.
Martín V, Valencia A, Agirre X, Cervera J, Jose‐Eneriz ES, Vilas‐Zornoza A, et al. Epigenetic regulation of the non‐canonical Wnt pathway in acute myeloid leukemia. Cancer Sci 2010;101(2):425-32. https://doi.org/10.1111/j.1349-7006.2009.01413.x.
Rao TP, Kühl M. An updated overview on Wnt signaling pathways: A prelude for more. Circ Res 2010;106(12):1798-806. https://doi.org/10.1161/CIRCRESAHA.110.219840.
Robbs BK, Cruz AL, Werneck MB, Mognol GP, Viola JP. Dual roles for NFAT transcription factor genes as oncogenes and tumor suppressors. Mol Cell Biol 2008;28(23):7168-81. https://doi.org/10.1128/MCB.00256-08.
Duque J, Fresno M, Iñiguez MA. Expression and function of the nuclear factor of activated T cells in colon carcinoma cells: Involvement in the regulation of cyclooxygenase-2. J Biol Chem 2005;280(10):8686-93. https://doi.org/10.1074/jbc.M413076200.
Viola J, Carvalho L, Fonseca B, Teixeira L. NFAT transcription factors: From cell cycle to tumor development. Braz J Med Biol Res 2005;38(3):335-44. https://doi.org/10.1590/S0100-879X2005000300003.
Yoeli-Lerner M, Yiu GK, Rabinovitz I, Erhardt P, Jauliac S, Toker A. Akt blocks breast cancer cell motility and invasion through the transcription factor NFAT. Mol Cell 2005;20(4):539-50. https://doi.org/10.1016/j.molcel.2005.10.033.
Buchholz M, Schatz A, Wagner M, Michl P, Linhart T, Adler G, et al. Overexpression of c‐myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J 2006;25(15):3714-24. https://doi.org/10.1038/sj.emboj.7601246.
Köenig A, Linhart T, Schlengemann K, Reutlinger K, Wegele J, Adler G, et al. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastroenterology 2010;138(3):1189-99. e1-2.
DOI: 10.1053/j.gastro.2009.10.045.
Tie X, Han S, Meng L, Wang Y, Wu A. NFAT1 is highly expressed in, and regulates the invasion of, glioblastoma multiforme cells. PLoS One 2013;8(6):e66008.
https://doi.org/10.1371/journal.pone.0066008.
Yiu GK, Toker A. NFAT induces breast cancer cell invasion by promoting the induction of cyclooxygenase-2. J Biol Chem 2006;281(18):12210-7. https://doi.org/10.1074/jbc.M600184200.
Jauliac S, López-Rodriguez C, Shaw LM, Brown LF, Rao A, Toker A. The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat Cell Biol 2002;4(7):540-4. https://doi.org/10.1038/ncb816.
Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature 1992;357:695-7. DOI: 10.1038/357695a0.
Shibasaki F, Price ER, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 1996;382(6589):370-3. https://doi.org/10.1038/382370a0.
Macian F. NFAT proteins: Key regulators of T-cell development and function. Nat Rev Immunol 2005;5(6):472-84. https://doi.org/10.1038/nri1632.
Daniel C, Gerlach K, Väth M, Neurath MF, Weigmann B. Nuclear factor of activated T cells - A transcription factor family as critical regulator in lung and colon cancer. Int J Cancer 2014;134(8):1767-75. https://doi.org/10.1002/ijc.28329.
Papi A, Rocchi P, Ferreri AM, Orlandi M. RXRγ and PPARγ ligands in combination to inhibit proliferation and invasiveness in colon cancer cells. Cancer Lett 2010;297(1):65-74. https://doi.org/10.1016/j.canlet.2010.04.026.
Sabatino L, Fucci A, Pancione M, Colantuoni V. PPARG epigenetic deregulation and its role in colorectal tumorigenesis. PPAR research 2012;2012. http://dx.doi.org/10.1155/2012/687492.
Houseknecht KL, Cole BM, Steele PJ. Peroxisome proliferator-activated receptor gamma (PPARγ) and its ligands: A review. Domest Anim Endocrinol 2002;22(1):1-23.
https://doi.org/10.1016/S0739-7240(01)00117-5.
Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: Complex stories. Nat Rev Cancer 2004;4(1):61-70. https://doi.org/10.1038/nrc1254.
Schweitzer A, Knauer SK, Stauber RH. Nuclear receptors in head and neck cancer: Current knowledge and perspectives. Int J Cancer 2010;126(4):801-9.
DOI: 10.1002/ijc.24968.
Lehrke M, Lazar MA. The many faces of PPARγ. Cell 2005;123(6):993-9. https://doi.org/10.1016/j.cell.2005.11.026.
Welch JS, Ricote M, Akiyama TE, Gonzalez FJ, Glass CK. PPARγ and PPARδ negatively regulate specific subsets of lipopolysaccharide and IFN-γ target genes in macrophages. Proc Natl Acad Sci U S A 2003;100(11):6712-7. https://doi.org/10.1073/pnas.1031789100.
Fajas L, Auboeuf D, Raspé E, Schoonjans K, Lefebvre AM, Saladin R, et al. The organization, promoter analysis, and expression of the human PPARγ gene. J Biol Chem 1997;272(30):18779-89. https://doi.org/10.1074/jbc.272.30.18779.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 2001;29(9):e45.
Orang AV, Safaralizadeh R, Hosseinpour Feizi M, Somi MH. Diagnostic relevance of overexpressed serine threonine tyrosine kinase/novel oncogene with kinase domain (STYK1/NOK) mRNA in colorectal cancer. Asian Pac J Cancer Prev 2014;15(16):6685-9. https://doi.org/10.7314/APJCP.2014.15.16.6685.
Hashemzadeh S, Arabzadeh AA, Estiar MA, Sakhinia M, Mesbahi N, Emrahi L, et al. Clinical utility of measuring expression levels of Stanniocalcin 2 in patients with colorectal cancer. Medical Oncol 2014;31(10):1-7.
https://doi.org/10.1007/s12032-014-0237-8.
Gerlach K, Daniel C, Lehr HA, Nikolaev A, Gerlach T, Atreya R, et al. Transcription factor NFATc2 controls the emergence of colon cancer associated with IL-6-dependent colitis. Cancer Res 2012;72(17):4340-50.
https://doi.org/10.1158/0008-5472.CAN-11-4155.
Qin L, Zhao D, Liu X, Nagy JA, Van Hoang M, Brown LF, et al. Down syndrome candidate region 1 isoform 1 mediates angiogenesis through the calcineurin-NFAT pathway. Mol Cancer Res 2006;4(11):811-20.
https://doi.org/10.1158/1541-7786.MCR-06-0126.
Baumgart S, Glesel E, Singh G, Chen NM, Reutlinger K, Zhang J, et al. Restricted heterochromatin formation links NFATc2 repressor activity with growth promotion in pancreatic cancer. Gastroenterology 2012;142(2):388-98. e1-7.
DOI: 10.1053/j.gastro.2011.11.001.
Maxeiner JH, Karwot R, Sauer K, Scholtes P, Boross I, Koslowski M, et al. A key regulatory role of the transcription factor NFATc2 in bronchial adenocarcinoma via CD8+ T lymphocytes. Cancer Res 2009;69(7):3069-76.
https://doi.org/10.1158/0008-5472.CAN-08-1678.
Panza A, Pazienza V, Ripoli M, Benegiamo G, Gentile A, Valvano MR, et al. Interplay between SOX9, β-catenin and PPARγ activation in colorectal cancer. Biochim Biophys Acta 2013;1833(8):1853-65. https://doi.org/10.1016/j.bbamcr.2013.04.004.
Chen LC, Hao CY, Chiu YS, Wong P, Melnick JS, Brotman M, et al. Alteration of gene expression in normal-appearing colon mucosa of APCmin mice and human cancer patients. Cancer Res 2004;64(10):3694-700. https://doi.org/10.1158/0008-5472.CAN-03-3264.
Yang L, Zhang H, Zhou ZG, Yan H, Adell G, Sun XF. Biological function and prognostic significance of peroxisome proliferator-activated receptor δ in rectal cancer. Clin Cancer Res 2011;17(11):3760-70. https://doi.org/10.1158/1078-0432.CCR-10-2779.
Dai Y, Qiao L, Chan KW, Yang M, Ye J, Ma J, et al. Peroxisome proliferator-activated receptor-γ contributes to the inhibitory effects of embelin on colon carcinogenesis. Cancer Res 2009;69(11):4776-83. https://doi.org/10.1158/0008-5472.CAN-08-4754.
Menendez JA. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: Molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta 2010;1801(3):381-91. https://doi.org/10.1016/j.bbalip.2009.09.005.
Ogino S, Shima K, Baba Y, Nosho K, Irahara N, Kure S, et al. Colorectal cancer expression of peroxisome proliferator-activated receptor γ (PPARG, PPAR gamma) is associated with good prognosis. Gastroenterology 2009;136(4):1242-50. https://doi.org/10.1053/j.gastro.2008.12.048.
Peri A, Cellai I, Benvenuti S, Luciani P, Baglioni S, Serio M. PPARgamma in neuroblastoma. PPAR Res 2008;2008:917815. DOI: 10.1155/2008/917815.
Downloads
Additional Files
Published
Issue
Section
Categories
How to Cite
Accepted 2017-02-24
Published 2017-08-20









