TNFSF14 and CD44 are overexpressed in glioblastoma and associated with immunosuppressive microenvironment
DOI:
https://doi.org/10.17305/bb.2025.11791Keywords:
Glioblastoma, GBM, cluster of differentiation 44, CD44, tumor necrosis factor superfamily member 14, TNFSF14, immune-checkpoints, tumor microenvironment, TMEAbstract
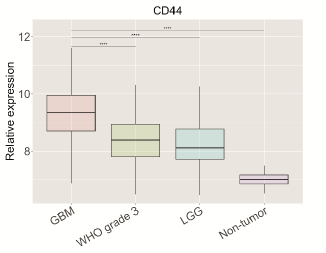
Glioblastoma (GBM) is one of the deadliest cancers, and the survival rate has remained low for decades. The aim of the study was the construction of the programmed death-ligand 1 (PD-L1) network, identification of its interactors and over-represented pathways, and analysis of the association between the identified genes and the immunosuppressive microenvironment of GBM. The PD-L1 network was constructed using Cytoscape and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). Over-representation analysis was performed on WebGestalt using Kyoto Encyclopedia of Genes and Genomes (KEGG), Protein ANalysis THrough Evolutionary Relationships (Panther), and Reactome Pathway Database (Reactome). Gene expression levels were examined in silico using three large datasets (The Cancer Genome Atlas (TCGA), Chinese Glioma Genome Atlas (CGGA), and Rembrandt), as well as with qPCR. The association between PD-L1 gene expression and immune cell infiltration was analyzed using the Tumor Immune Estimation Resource (TIMER 2.0) online tool. Cluster of differentiation 44 (CD44) and tumor necrosis factor superfamily member 14 (TNFSF14) were found to be significantly overexpressed in GBM compared to lower-grade glioma (LGG) and normal brain tissue. Their overexpression was associated with worse overall survival and demonstrated a strong ability to differentiate between GBM and reference brain tissue. Notably, CD44 and TNFSF14 were linked to the mesenchymal subtype of GBM and positively correlated with the presence of regulatory T cells, resting natural killer (NK) cells, and PD-L1 expression. Our findings highlight the overexpression of CD44 and TNFSF14 in GBM and their potential involvement in creating an immunosuppressive microenvironment. Unraveling the PD-L1 interaction network and its associated pathways offers the potential not only to identify novel biomarkers for GBM prognosis but also to pinpoint alternative therapeutic targets that could be more effective in overcoming the immunosuppressive hurdles inherent in GBM treatment.
Citations
Downloads
Downloads
Published
License
Copyright (c) 2025 Alja Zottel, Neja Šamec, Ivana Jovčevska

This work is licensed under a Creative Commons Attribution 4.0 International License.









