Cancer metastasis - tricks of the trade
DOI:
https://doi.org/10.17305/bjbms.2017.1908Keywords:
Cancer, metastasis, tumor cell infiltration, disseminated tumor cells, circulating tumor cellsAbstract
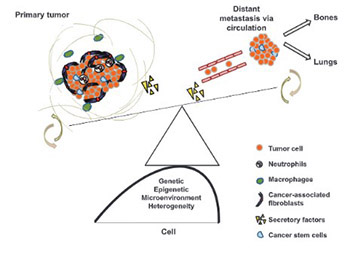
Decades of cancer research have unraveled genetic, epigenetic and molecular pathways leading to plausible therapeutic targets; many of which hold great promise in improving clinical outcomes. Metastatic tumors become evident early on and are one of the major causes of cancer-related fatalities worldwide. This review depicts the sequential events of cancer metastasis. Genetic and epigenetic heterogeneity influences local tumor cell invasion, intravasation, survival in circulation, extravasation and colonization to distant sites. Each sequential event is associated with heterogeneous tumor microenvironment, gain of competence, unique population of cancer stem cells (CSCs), circulatory pathway, compatible niche and immune system support. A tight regulation of metastasis-promoting mechanisms and, in parallel, evading inhibitory mechanisms contribute to the severity and site of metastasis. A comprehensive understanding of tumor cell fate as an individual entity, as well as in combination with different promoting factors and associated molecular mechanisms, is anticipated in the coming years. This will enable scientists to depict design strategies for targeted cancer therapies.
Citations
Downloads
References
Christofori G. New signals from the invasive front. Nature 2006;441(7092):444-50. https://doi.org/10.1038/nature04872.
Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell 2006;127(4):679-95. https://doi.org/10.1016/j.cell.2006.11.001.
Steeg PS. Tumor metastasis: Mechanistic insights and clinical challenges. Nat Med 2006;12(8):895-904. https://doi.org/10.1038/nm1469.
Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011;144(5):646-74. https://doi.org/10.1016/j.cell.2011.02.013.
McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 2014;16(8):717-27. https://doi.org/10.1038/ncb3015.
Nguyen DX, Bos PD, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer 2009;9(4):274-84. https://doi.org/10.1038/nrc2622.
Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer 2002;2(8):563-72. https://doi.org/10.1038/nrc865.
Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450(7173):1235-9. https://doi.org/10.1038/nature06385.
Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med 2005;353(8):793-802. https://doi.org/10.1056/NEJMoa050434.
Wong CW, Lee A, Shientag L, Yu J, Dong Y, Kao G, et al. Apoptosis: An early event in metastatic inefficiency. Cancer Res 2001;61(1):333-8.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature 2005;436(7050):518-24. https://doi.org/10.1038/nature03799.
Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature 2011;481(7379):85-9. https://doi.org/10.1038/nature10694.
Capasso LL. Antiquity of cancer. Int J Cancer 2005;113(1):2-13. https://doi.org/10.1002/ijc.20610.
Paget S. The distribution of secondary growths in cancer of the breast 1889. Cancer Metastasis Rev 1989;8(2):98-101.
James Ewing AM. Neoplastic Diseases: A Treatise on Tumours. 3rd edition, Vol. 8. Philadelphia, London: W. B. Saunders Co., Ltd.; 1928.
Wan L, Pantel K, Kang Y. Tumor metastasis: Moving new biological insights into the clinic. Nat Med 2013;19(11):1450-64. https://doi.org/10.1038/nm.3391.
Talmadge JE, Fidler IJ. AACR centennial series: The biology of cancer metastasis: Historical perspective. Cancer Res 2010;70(14):5649-69.
https://doi.org/10.1158/0008-5472.CAN-10-1040.
Fidler IJ. The pathogenesis of cancer metastasis: The seed and soil hypothesis revisited. Nat Rev Cancer 2003;3(6):453-8. https://doi.org/10.1038/nrc1098.
Hanahan D, Coussens LM. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012;21(3):309-22. https://doi.org/10.1016/j.ccr.2012.02.022.
Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med 2013;19(11):1423-37. https://doi.org/10.1038/nm.3394.
Muller C, Holtschmidt J, Auer M, Heitzer E, Lamszus K, Schulte A, et al. Hematogenous dissemination of glioblastoma multiforme. Sci Transl Med 2014;6(247):247ra101. https://doi.org/10.1126/scitranslmed.3009095.
Sullivan JP, Nahed BV, Madden MW, Oliveira SM, Springer S, Bhere D, et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov 2014;4(11):1299-309. https://doi.org/10.1158/2159-8290.CD-14-0471.
Pietschmann S, von Bueren AO, Kerber MJ, Baumert BG, Kortmann RD, Müller K. An individual patient data meta-analysis on characteristics, treatments and outcomes of glioblastoma/gliosarcoma patients with metastases outside of the central nervous system. PLoS One 2015;10(4):e0121592. https://doi.org/10.1371/journal.pone.0121592.
Mohme M, Riethdorf S, Pantel K. Circulating and disseminated tumour cells - mechanisms of immune surveillance and escape. Nat Rev Clin Oncol 2016. https://doi.org/10.1038/nrclinonc.2016.144.
Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med 2015;21(8):846-53. https://doi.org/10.1038/nm.3915.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science 2013;339(6127):1546-58. https://doi.org/10.1126/science.1235122.
Stupack DG, Teitz T, Potter MD, Mikolon D, Houghton PJ, Kidd VJ, et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature 2006;439(7072):95-9. https://doi.org/10.1038/nature04323.
Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res 2008;68(9):3108-14. https://doi.org/10.1158/0008-5472.CAN-07-5644.
Hunter K. Host genetics influence tumour metastasis. Nat Rev Cancer 2006;6(2):141-6. https://doi.org/10.1038/nrc1803.
Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363(18):1693-703. https://doi.org/10.1056/NEJMoa1006448.
Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344(11):783-92. https://doi.org/10.1056/NEJM200103153441101.
Staller P, Sulitkova J, Lisztwan J, Moch H, Oakeley EJ, Krek W. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature 2003;425(6955):307-11. https://doi.org/10.1038/nature01874.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003;3(6):537-49. https://doi.org/10.1016/S1535-6108(03)00132-6.
Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol 2009;11(11):1287-96. https://doi.org/10.1038/ncb1973.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139(5):871-90. https://doi.org/10.1016/j.cell.2009.11.007.
De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013;13(2):97-110. https://doi.org/10.1038/nrc3447.
Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013;19(11):1438-49. https://doi.org/10.1038/nm.3336.
Bednarz-Knoll N, Alix-Panabières C, Pantel K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev 2012;31(3-4):673-87. https://doi.org/10.1007/s10555-012-9370-z.
Yokobori T, Iinuma H, Shimamura T, Imoto S, Sugimachi K, Ishii H, et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res 2013;73(7):2059-69. https://doi.org/10.1158/0008-5472.CAN-12-0326.
Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010;141(1):52-67. https://doi.org/10.1016/j.cell.2010.03.015.
Weis SM, Cheresh DA. Tumor angiogenesis: Molecular pathways and therapeutic targets. Nat Med 2011;17(11):1359-70. https://doi.org/10.1038/nm.2537.
Bandyopadhyay S, Zhan R, Chaudhuri A, Watabe M, Pai SK, Hirota S, et al. Interaction of KAI1 on tumor cells with DARC on vascular endothelium leads to metastasis suppression. Nat Med 2006;12(8):933-8. https://doi.org/10.1038/nm1444.
Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011;147(2):275-92. https://doi.org/10.1016/j.cell.2011.09.024.
Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell 2008;133(1):66-77. https://doi.org/10.1016/j.cell.2008.01.046.
Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 2007;446(7137):765-70. https://doi.org/10.1038/nature05760.
Karpatkin S, Pearlstein E. Role of platelets in tumor cell metastases. Ann Intern Med 1981;95(5):636-41. https://doi.org/10.7326/0003-4819-95-5-636.
Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer 2004;4(6):448-56. https://doi.org/10.1038/nrc1370.
Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci U S A 2003;100(13):7737-42. https://doi.org/10.1073/pnas.1331931100.
Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002;2(6):442-54. https://doi.org/10.1038/nrc822.
Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat Rev Cancer 2004;4(2):118-32. https://doi.org/10.1038/nrc1276.
Richards FM, McKee SA, Rajpar MH, Cole TR, Evans DG, Jankowski JA, et al. Germline E-cadherin gene (CDH1) mutations predispose to familial gastric cancer and colorectal cancer. Hum Mol Genet 1999;8(4):607-10. https://doi.org/10.1093/hmg/8.4.607.
Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006;439(7074):353-7. https://doi.org/10.1038/nature04296.
Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 2004;117(7):927-39. https://doi.org/10.1016/j.cell.2004.06.006.
Calbo J, van Montfort E, Proost N, van Drunen E, Beverloo HB, Meuwissen R, et al. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011;19(2):244-56. https://doi.org/10.1016/j.ccr.2010.12.021.
Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell 2006;124(1):207-19. https://doi.org/10.1016/j.cell.2005.10.043.
Muller PA, Vousden KH, Norman JC. P53 and its mutants in tumor cell migration and invasion. J Cell Biol 2011;192(2):209-18. DOI: 10.1083/jcb.201009059.
Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, et al. P53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011;13(3):317-23. https://doi.org/10.1038/ncb2173.
Godar S, Ince TA, Bell GW, Feldser D, Donaher JL, Bergh J, et al. Growth-inhibitory and tumor-suppressive functions of p53 depend on its repression of CD44 expression. Cell 2008;134(1):62-73. https://doi.org/10.1016/j.cell.2008.06.006.
Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov 2014;4(4):405-14. https://doi.org/10.1158/2159-8290.CD-13-0136.
Douma S, Van Laar T, Zevenhoven J, Meuwissen R, Van Garderen E, Peeper DS. Suppression of anoikis and induction of metastasis by the neurotrophic receptor TrkB. Nature 2004;430(7003):1034-9. https://doi.org/10.1038/nature02765.
Tabassum DP, Polyak K. Tumorigenesis: It takes a village. Nat Rev Cancer 2015;15(8):473-83. https://doi.org/10.1038/nrc3971.
Marusyk A, Tabassum DP, Altrock PM, Almendro V, Michor F, Polyak K. Non-cell-autonomous driving of tumour growth supports sub-clonal heterogeneity. Nature 2014;514(7520):54-8. https://doi.org/10.1038/nature13556.
Cleary AS, Leonard TL, Gestl SA, Gunther EJ. Tumour cell heterogeneity maintained by cooperating subclones in Wnt-driven mammary cancers. Nature 2014;508(7494):113-7. https://doi.org/10.1038/nature13187.
Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158(5):1110-22. https://doi.org/10.1016/j.cell.2014.07.013.
Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JM, Papaemmanuil E, et al. The evolutionary history of lethal metastatic prostate cancer. Nature 2015;520(7547):353-7. https://doi.org/10.1038/nature14347.
Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009;459(7249):1005-9. https://doi.org/10.1038/nature08021.
Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 2011;20(5):576-90. https://doi.org/10.1016/j.ccr.2011.09.009.
Piskounova E, Agathocleous M, Murphy MM, Hu Z, Huddlestun SE, Zhao Z, et al. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature 2015;527(7577):186-91. https://doi.org/10.1038/nature15726.
Chen Q, Zhang XH, Massagué J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell 2011;20(4):538-49. https://doi.org/10.1016/j.ccr.2011.08.025.
Denève E, Riethdorf S, Ramos J, Nocca D, Coffy A, Daurès JP, et al. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin Chem 2013;59(9):1384-92. https://doi.org/10.1373/clinchem.2013.202846.
Al-Mehdi AB, Tozawa K, Fisher AB, Shientag L, Lee A, Muschel RJ. Intravascular origin of metastasis from the proliferation of endothelium-attached tumor cells: A new model for metastasis. Nat Med 2000;6(1):100-2. https://doi.org/10.1038/71429.
Budczies J, von Winterfeld M, Klauschen F, Bockmayr M, Lennerz JK, Denkert C, et al. The landscape of metastatic progression patterns across major human cancers. Oncotarget 2015;6(1):570-83. DOI: 10.18632/oncotarget.2677.
Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res 2007;100(2):158-73. https://doi.org/10.1161/01.RES.0000255691.76142.4a.
Klein CA. Framework models of tumor dormancy from patient-derived observations. Curr Opin Genet Dev 2011;21(1):42-9. https://doi.org/10.1016/j.gde.2010.10.011.
Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007;7(11):834-46. https://doi.org/10.1038/nrc2256.
Pantel K, Alix-Panabières C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol 2009;6(6):339-51. https://doi.org/10.1038/nrclinonc.2009.44.
Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer 2009;9(4):302-12. https://doi.org/10.1038/nrc2627.
Jo H, Jia Y, Subramanian KK, Hattori H, Luo HR. Cancer cell-derived clusterin modulates the phosphatidylinositol 3'-kinase-Akt pathway through attenuation of insulin-like growth factor 1 during serum deprivation. Mol Cell Biol 2008;28(13):4285-99. https://doi.org/10.1128/MCB.01240-07.
Deng X, Ewton DZ, Friedman E. Mirk/Dyrk1B maintains the viability of quiescent pancreatic cancer cells by reducing levels of reactive oxygen species. Cancer Res 2009;69(8):3317-24. https://doi.org/10.1158/0008-5472.CAN-08-2903.
Ewton DZ, Hu J, Vilenchik M, Deng X, Luk KC, Polonskaia A, et al. Inactivation of mirk/dyrk1b kinase targets quiescent pancreatic cancer cells. Mol Cancer Ther 2011;10(11):2104-14. https://doi.org/10.1158/1535-7163.MCT-11-0498.
Murrell DH, Foster PJ, Chambers AF. Brain metastases from breast cancer: Lessons from experimental magnetic resonance imaging studies and clinical implications. J Mol Med (Berl) 2014;92(1):5-12. https://doi.org/10.1007/s00109-013-1108-z.
Heyn C, Ronald JA, Ramadan SS, Snir JA, Barry AM, MacKenzie LT, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med 2006;56(5):1001-10. https://doi.org/10.1002/mrm.21029.
Bragado P, Estrada Y, Parikh F, Krause S, Capobianco C, Farina HG, et al. TGF-ß2 dictates disseminated tumour cell fate in target organs through TGF-ß-RIII and p38a/ß signalling. Nat Cell Biol 2013;15(11):1351-61. https://doi.org/10.1038/ncb2861.
Aguirre-Ghiso JA, Bragado P, Sosa MS. Metastasis awakening: Targeting dormant cancer. Nat Med 2013;19(3):276-7. https://doi.org/10.1038/nm.3120.
Shiozawa Y, Havens AM, Pienta KJ, Taichman RS. The bone marrow niche: Habitat to hematopoietic and mesenchymal stem cells, and unwitting host to molecular parasites. Leukemia 2008;22(5):941-50. https://doi.org/10.1038/leu.2008.48.
Joseph J, Shiozawa Y, Jung Y, Kim JK, Pedersen E, Mishra A, et al. Disseminated prostate cancer cells can instruct hematopoietic stem and progenitor cells to regulate bone phenotype. Mol Cancer Res 2012;10(3):282-92.
https://doi.org/10.1158/1541-7786.MCR-11-0404.
Gao H, Chakraborty G, Lee-Lim AP, Mo Q, Decker M, Vonica A, et al. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell 2012;150(4):764-79. https://doi.org/10.1016/j.cell.2012.06.035.
Lu X, Mu E, Wei Y, Riethdorf S, Yang Q, Yuan M, et al. VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging a4ß1-positive osteoclast progenitors. Cancer Cell 2011;20(6):701-14. https://doi.org/10.1016/j.ccr.2011.11.002.
Barkan D, El Touny LH, Michalowski AM, Smith JA, Chu I, Davis AS, et al. Metastatic growth from dormant cells induced by a col-I-enriched fibrotic environment. Cancer Res 2010;70(14):5706-16. https://doi.org/10.1158/0008-5472.CAN-09-2356.
Ranganathan AC, Adam AP, Zhang L, Aguirre-Ghiso JA. Tumor cell dormancy induced by p38SAPK and ER-stress signaling: An adaptive advantage for metastatic cells? Cancer Biol Ther 2006;5(7):729-35. https://doi.org/10.4161/cbt.5.7.2968.
Sosa MS, Avivar-Valderas A, Bragado P, Wen HC, Aguirre-Ghiso JA. ERK1/2 and p38a/ß signaling in tumor cell quiescence: Opportunities to control dormant residual disease. Clin Cancer Res 2011;17(18):5850-7.
https://doi.org/10.1158/1078-0432.CCR-10-2574.
Clevers H. The cancer stem cell: Premises, promises and challenges. Nat Med 2011;17(3):313-9. https://doi.org/10.1038/nm.2304.
Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell 2008;132(4):598-611. https://doi.org/10.1016/j.cell.2008.01.038.
Hsu YC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med 2014;20(8):847-56. https://doi.org/10.1038/nm.3643.
Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, et al. Lineage tracing reveals Lgr5 stem cell activity in mouse intestinal adenomas. Science 2012;337(6095):730-5. https://doi.org/10.1126/science.1224676.
Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature 2012;488(7412):527-30. https://doi.org/10.1038/nature11344.
Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov 2014;13(7):497-512. https://doi.org/10.1038/nrd4253.
Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011;145(6):926-40. https://doi.org/10.1016/j.cell.2011.04.029.
Oskarsson T, Batlle E, Massagué J. Metastatic stem cells: Sources, niches, and vital pathways. Cell Stem Cell 2014;14(3):306-21. https://doi.org/10.1016/j.stem.2014.02.002.
Shiozawa Y, Pedersen EA, Havens AM, Jung Y, Mishra A, Joseph J, et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest 2011;121(4):1298-312. https://doi.org/10.1172/JCI43414.
Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007;1(3):313-23. https://doi.org/10.1016/j.stem.2007.06.002.
Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature 2016;529(7586):298-306. https://doi.org/10.1038/nature17038.
Lee YT. Patterns of metastasis and natural courses of breast carcinoma. Cancer Metastasis Rev 1985;4(2):153-72. https://doi.org/10.1007/BF00050693.
Johansson JE, Andrén O, Andersson SO, Dickman PW, Holmberg L, Magnuson A, et al. Natural history of early, localized prostate cancer. JAMA 2004;291(22):2713-9. https://doi.org/10.1001/jama.291.22.2713.
Feld R, Rubinstein LV, Weisenberger TH. Sites of recurrence in resected stage I non-small-cell lung cancer: A guide for future studies. J Clin Oncol 1984;2(12):1352-8. DOI: 10.1200/JCO.1984.2.12.1352.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990;61(5):759-67. https://doi.org/10.1016/0092-8674(90)90186-I.
Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, et al. Metastatic patterns in adenocarcinoma. Cancer 2006;106(7):1624-33. https://doi.org/10.1002/cncr.21778.
Soucek L, Lawlor ER, Soto D, Shchors K, Swigart LB, Evan GI. Mast cells are required for angiogenesis and macroscopic expansion of Myc-induced pancreatic islet tumors. Nat Med 2007;13(10):1211-8. https://doi.org/10.1038/nm1649.
Sparmann A, Bar-Sagi D. Ras-induced interleukin-8 expression plays a critical role in tumor growth and angiogenesis. Cancer Cell 2004;6(5):447-58. https://doi.org/10.1016/j.ccr.2004.09.028.
Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature 2008;454(7203):436-44. https://doi.org/10.1038/nature07205.
Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004;431(7007):461-6. https://doi.org/10.1038/nature02924.
Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med 2004;10(1):48-54. https://doi.org/10.1038/nm976.
Zhou C, Liu J, Tang Y, Liang X. Inflammation linking EMT and cancer stem cells. Oral Oncol 2012;48(11):1068-75. https://doi.org/10.1016/j.oraloncology.2012.06.005.
Blackwell TS, Christman JW. The role of nuclear factor-kappa B in cytokine gene regulation. Am J Respir Cell Mol Biol 1997;17(1):3-9. https://doi.org/10.1165/ajrcmb.17.1.f132.
Coussens LM, Raymond WW, Bergers G, Laig-Webster M, Behrendtsen O, Werb Z, et al. Inflammatory mast cells up-regulate angiogenesis during squamous epithelial carcinogenesis. Genes Dev 1999;13(11):1382-97. https://doi.org/10.1101/gad.13.11.1382.
Oppenheimer SB. Cellular basis of cancer metastasis: A review of fundamentals and new advances. Acta Histochem 2006;108(5):327-34. https://doi.org/10.1016/j.acthis.2006.03.008.
Lin KY, Lu D, Hung CF, Peng S, Huang L, Jie C, et al. Ectopic expression of vascular cell adhesion molecule-1 as a new mechanism for tumor immune evasion. Cancer Res 2007;67(4):1832-41. https://doi.org/10.1158/0008-5472.CAN-06-3014.
Yan HH, Pickup M, Pang Y, Gorska AE, Li Z, Chytil A, et al. Gr-1 CD11b myeloid cells tip the balance of immune protection to tumor promotion in the premetastatic lung. Cancer Res 2010;70(15):6139-49. https://doi.org/10.1158/0008-5472.CAN-10-0706.
Lee JJ, Murphy GF, Lian CG. Melanoma epigenetics: Novel mechanisms, markers, and medicines. Lab Invest 2014;94(8):822-38. https://doi.org/10.1038/labinvest.2014.87.
Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 2009;139(4):693-706. https://doi.org/10.1016/j.cell.2009.10.014.
Brower V. Epigenetics: Unravelling the cancer code. Nature 2011;471(7339):S12-3. https://doi.org/10.1038/471S12a.
Ju HX, An B, Okamoto Y, Shinjo K, Kanemitsu Y, Komori K, et al. Distinct profiles of epigenetic evolution between colorectal cancers with and without metastasis. Am J Pathol 2011;178(4):1835-46. https://doi.org/10.1016/j.ajpath.2010.12.045.
Muthusamy V, Premi S, Soper C, Platt J, Bosenberg M. The hematopoietic stem cell regulatory gene latexin has tumor-suppressive properties in malignant melanoma. J Invest Dermatol 2013;133(7):1827-33. https://doi.org/10.1038/jid.2013.48.
Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005;37(4):391-400. https://doi.org/10.1038/ng1531.
Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 1997;89(3):349-56. https://doi.org/10.1016/S0092-8674(00)80215-9.
Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007;26(37):5420-32. https://doi.org/10.1038/sj.onc.1210610.
Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 2004;59(2):177-89. https://doi.org/10.1002/pros.20022.
Song J, Noh JH, Lee JH, Eun JW, Ahn YM, Kim SY, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS 2005;113(4):264-8. https://doi.org/10.1111/j.1600-0463.2005.apm_04.x.
Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol 2004;24(1):306-19. https://doi.org/10.1128/MCB.24.1.306-319.2004.
Serman L, Nikuseva Martic T, Serman A, Vranic S. Epigenetic alterations of the Wnt signaling pathway in cancer: A mini review. Bosn J Basic Med Sci 2014;14(4):191-4. https://doi.org/10.17305/bjbms.2014.4.205.
Batra S, Shi Y, Kuchenbecker KM, He B, Reguart N, Mikami I, et al. Wnt inhibitory factor-1, a Wnt antagonist, is silenced by promoter hypermethylation in malignant pleural mesothelioma. Biochem Biophys Res Commun 2006;342(4):1228-32. https://doi.org/10.1016/j.bbrc.2006.02.084.
Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004;36(4):417-22. https://doi.org/10.1038/ng1330.
Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, Cameron JL, et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res 2003;63(13):3735-42.
O'Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: Role in therapy response, resistance, and clinical outcome. Clin Cancer Res 2015;21(2):249-57. https://doi.org/10.1158/1078-0432.CCR-14-0990.
Haeno H, Gonen M, Davis MB, Herman JM, Iacobuzio-Donahue CA, Michor F. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell 2012;148(1-2):362-75. https://doi.org/10.1016/j.cell.2011.11.060.
Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods 2014;11(4):417-22. https://doi.org/10.1038/nmeth.2869.
Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, et al. Multiplexed ion beam imaging of human breast tumors. Nat Med 2014;20(4):436-42. https://doi.org/10.1038/nm.3488.
Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 2015;10(3):442-58. https://doi.org/10.1038/nprot.2014.191.
Murphy PJ, Cipriany BR, Wallin CB, Ju CY, Szeto K, Hagarman JA, et al. Single-molecule analysis of combinatorial epigenomic states in normal and tumor cells. Proc Natl Acad Sci U S A 2013;110(19):7772-7. https://doi.org/10.1073/pnas.1218495110.
Downloads
Additional Files
Published
How to Cite
Accepted 2017-01-22
Published 2017-08-20









